Technologies
- Fee-for-service Assays
- Single-cell Epigenomics Platform
- The Epigenomics Platform
- Spatial Genomics & Epigenomics Platform
The Spatial Genomics & Epigenomics Platform specialize in advanced epigenomic technologies designed to explore gene expression in healthy tissue, uncover spatial patterns in disease, or identify potential therapeutic targets.
Our platform and functionally analyzes both cis- and trans-regulatory elements of mammalian genomes. Cis elements, such as promoters and enhancers, and trans-acting factors, such as transcription factors and other proteins, play pivotal roles in gene regulation. By examining both types of regulatory elements in tandem, this platform provides a more comprehensive understanding of how the epigenome governs cellular processes.
A unique and powerful feature of this platform is its focus on integrating spatial information with functional genomics. While traditional genomic methods often focus on bulk populations of cells, losing critical information about the individual cellular environment, the Spatial Genomics & Epigenomics Platform develops and adopts advanced imaging techniques to retain and analyze the spatial organization of cells. This approach enables researchers to investigate how epigenetic modifications and gene expression are influenced by the spatial and structural context of cells, tissues, and organs. Understanding the spatial organization of cells within their native tissue environment is essential for uncovering the epigenomic mechanisms that drive cell identity, behavior, and function. In disease states, such as cancer or neurological disorders, spatial disruptions at the cellular and epigenomic levels often lead to dysregulation of normal cellular functions.
Through a combination of state-of-the-art imaging and integrated computational analysis with sequencing, this platform equips researchers with the tools needed to decode the complex epigenomic landscapes that define cellular identity and function. By integrating spatial context into these analyses, it offers an unparalleled perspective on how the epigenome shapes health and disease.
MERFISH
Multiplexed Error Robust Fluorescence In situ Hybridization (MERFISH) is an imaging method capable of simultaneously measuring the copy number and spatial distribution of hundreds to thousands of RNA species in single cells. The Center collaborate closely with the pioneering laboratory of Xiaowei Zhuang at Harvard University to bring this groundbreaking technology to San Diego researchers. By leveraging the expertise of Dr. Zhuang’s lab, a leader in spatial genomics and super-resolution imaging, the Center is positioned to offer unparalleled access to transformative techniques that provide deep insights into gene expression and cellular function.
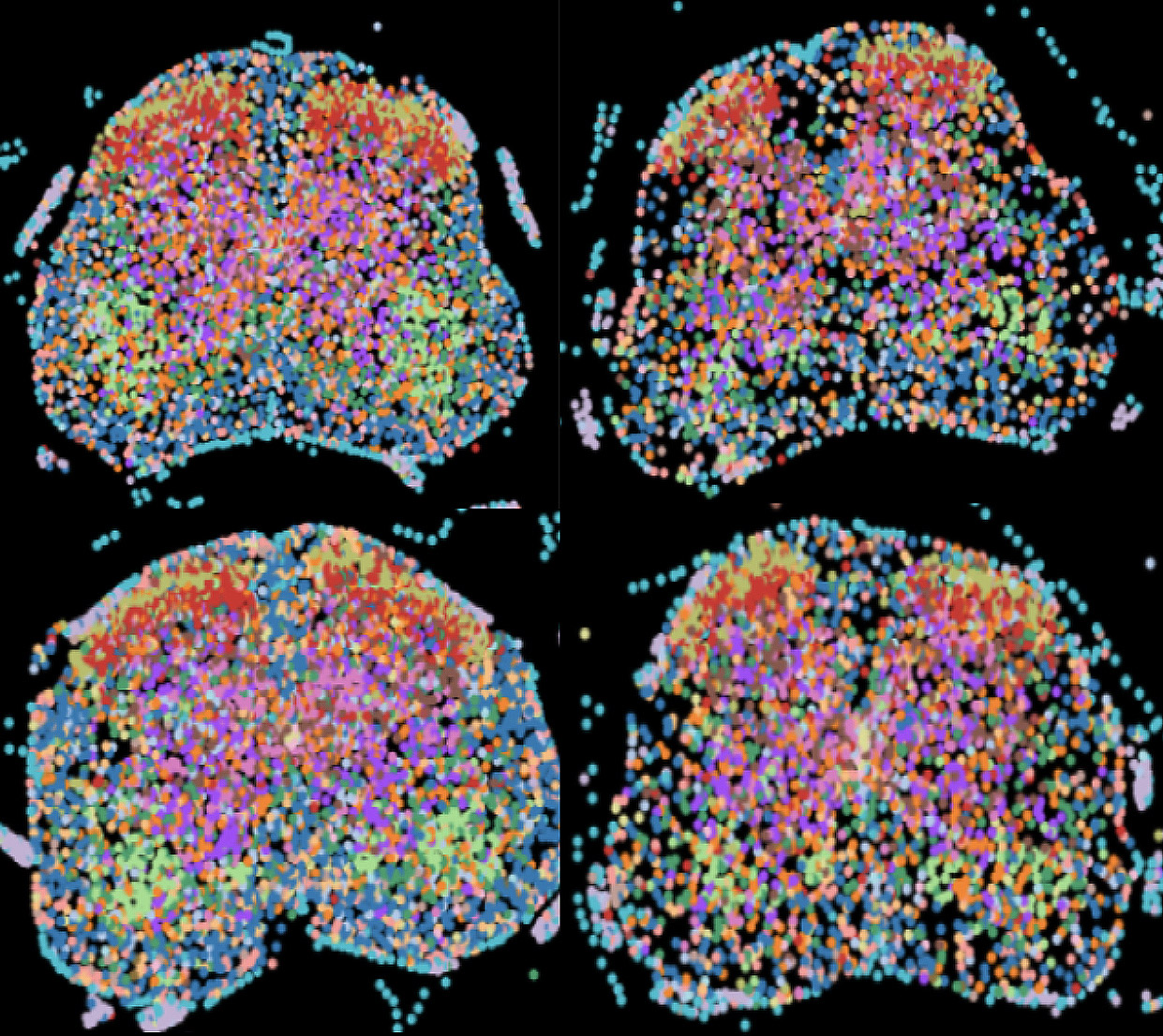 Currently, the Center offers the ability to image transcripts of 300, 500, and up to 3,000 genes with remarkable precision. This technology allows researchers to precisely quantify the copy numbers of these transcripts while simultaneously mapping their 3D localization within the cellular environment.
Currently, the Center offers the ability to image transcripts of 300, 500, and up to 3,000 genes with remarkable precision. This technology allows researchers to precisely quantify the copy numbers of these transcripts while simultaneously mapping their 3D localization within the cellular environment.
The spatial context provided by these methods is invaluable for understanding how genes are regulated and expressed in situ, shedding light on how cells function within complex tissues and organ systems.This can be transformative for studies in fields such as developmental biology, neurobiology, immunology, and cancer biology, where the spatial arrangement of cells and their gene expression can reveal new insights into cellular interactions, differentiation pathways, and disease mechanisms.
In addition to the cutting-edge technologies, the Center offers hands-on support and guidance, ensuring that researchers can seamlessly integrate these techniques into their studies.
Example Publication: https://www.nature.com/articles/s41586-024-07171-z
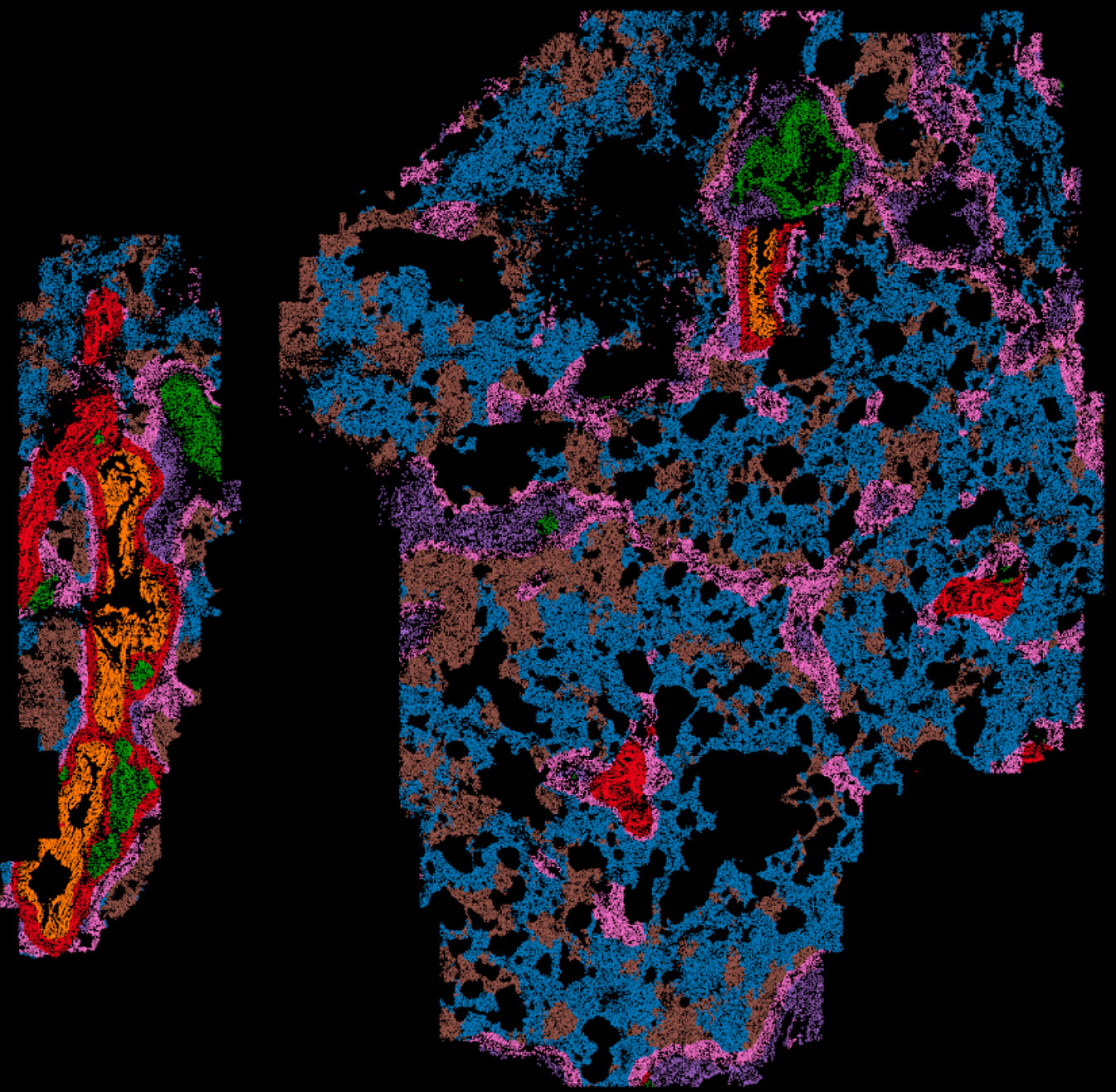 MERSCOPE
MERSCOPE
With multiple MERSCOPE instruments available, researchers can perform high-resolution spatial mapping of up to 960 RNA targets, providing subcellular resolution down to ≤100nm. This enables precise localization and quantification of RNA transcripts within their native cellular environment. Our MERSCOPE platform features built-in machine learning algorithms for on-instrument cell segmentation, enhancing the accuracy of data analysis. By integrating nuclei staining, total mRNA abundance (poly-A), and cell boundary staining, the system delivers highly accurate cell segmentation and spatial transcriptomic profiling.
In addition to custom panels, the Center offers a range of predesigned panels to streamline your research. These include Human PanCancer Pathways, Human Immuno-Oncology, and Mouse PanNeuro, all of which are compatible with both frozen and FFPE (formalin-fixed, paraffin-embedded) tissue samples, ensuring flexibility across diverse research applications.
Xenium
The Center offers high-throughput RNA Fluorescence In Situ Hybridization (FISH) through the cutting-edge Xenium Analyzer from 10X Genomics, enabling precise spatial mapping of RNA targets at subcellular resolution (~0.2 microns/pixel). With this powerful technology, researchers can map 480 custom RNA targets or select from 5,000 predesigned RNA targets, providing unparalleled insights into gene expression patterns within the native tissue context.
Each Xenium instrument run can profile two slides simultaneously, covering a total area of 5.76 cm², making it an ideal solution for large-scale studies. The instrument uses advanced machine learning to achieve on-instrument cell segmentation by combining nuclei staining, interior-RNA and protein abundance, and cell boundary staining, ensuring high accuracy in defining cellular boundaries and transcript localization.
One of the unique features of the Xenium platform is the ability to perform H&E (hematoxylin and eosin) or IF (immunofluorescence) staining on the same tissue slide following a Xenium run. This enables researchers to overlay traditional pathological analysis with high-resolution gene expression data, delivering a comprehensive view of both tissue morphology and molecular profiling.
The Center also provides a range of predesigned panels tailored to specific research needs, including Human Brain, Human Breast, Human Lung, Human Immuno-Oncology, Human Colon, Human Skin, Human Multi-Tissue, Human 5000 Gene, Mouse Brain, Mouse Multi-Tissue, and Mouse 5000 Gene panels. The platform is compatible with both frozen and FFPE (formalin-fixed, paraffin-embedded) samples, offering flexibility across various sample types.
Super-Resolution Chromatin Tracing
Proper organization of chromatin architecture has been increasingly recognized as a determining factor in regulating gene expression during normal cell cycle and development. Next generation sequencing-based technology, such as single nuclei Hi-C, has provided unprecedented information about chromatin architecture at single cell resolution. However, the application of this methodology has been limited by lack of spatial information, coupled with high-cost but low-throughput.
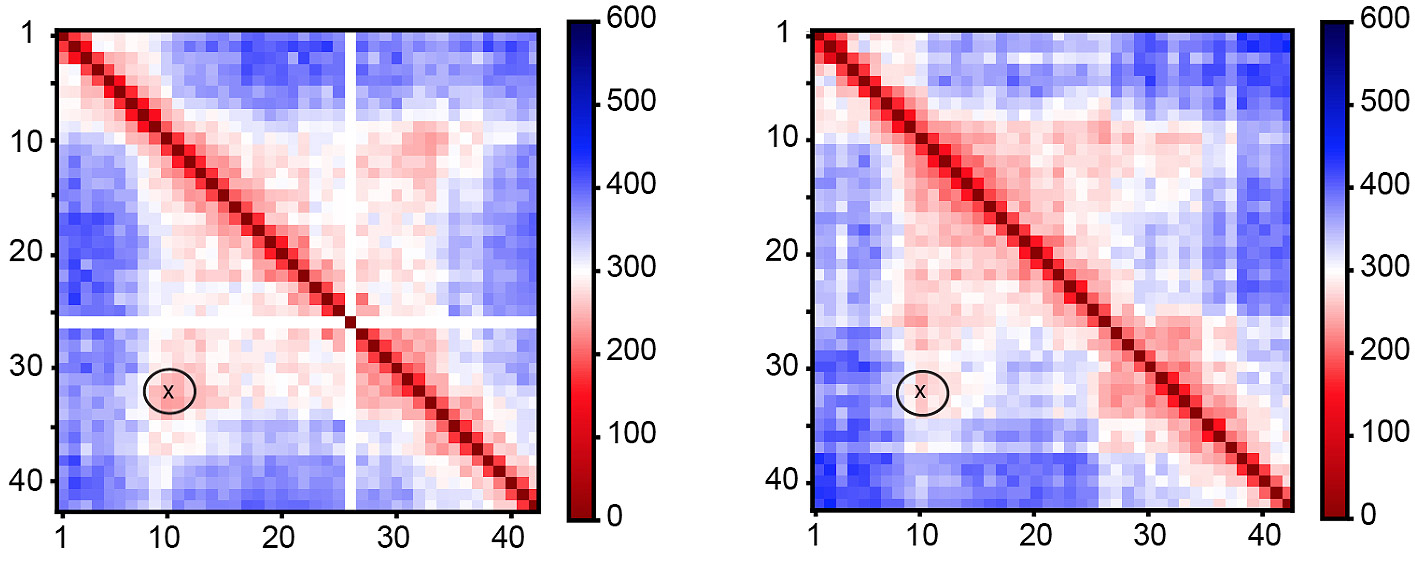 A newly developed imaging technology has allowed for visualization of chromatin architecture with hybridization of multiplexed FISH probes. Such high-coverage oligopaint FISH has revealed higher order chromatin structure that are highly consistent with that from Hi-C experiments. With multiple custom-built imaging systems, we are able to visualize individual chromatin 3D architecture from 2kb-ultra high resolution to genome-scale using sequential, multiplexed, high-throughput hybridization and imaging of oligopaint probes.
A newly developed imaging technology has allowed for visualization of chromatin architecture with hybridization of multiplexed FISH probes. Such high-coverage oligopaint FISH has revealed higher order chromatin structure that are highly consistent with that from Hi-C experiments. With multiple custom-built imaging systems, we are able to visualize individual chromatin 3D architecture from 2kb-ultra high resolution to genome-scale using sequential, multiplexed, high-throughput hybridization and imaging of oligopaint probes.
Example Publication: https://pubmed.ncbi.nlm.nih.gov/34002095/