Unveiling Chromatin Architecture with Droplet Hi-C: A Breakthrough in Single-Cell Analysis
New technique for mapping the brain and decoding cancer has also shed light on the dynamics of “extra-chromosomal” DNA
Imagine trying to fit a two-meter-long string inside the equivalent of a microscopic box, without the string becoming tangled. That’s essentially what’s happening with human DNA inside our cells. DNA, the source of our genetic code, is packaged into something called chromatin, allowing it to be tightly folded into a small space. But this 3D folding isn't random; it's organized in specific ways that help control which genes get turned on or off.
To understand diseases like cancer and neurological disorders, scientists must map how DNA folds and organizes itself inside cells. This organization plays a key role in gene regulation, which affects how cells behave—whether they're healthy or diseased. Scientists use a technique called Hi-C to map these DNA interactions, helping them visualize how the genome is folded in 3D space.
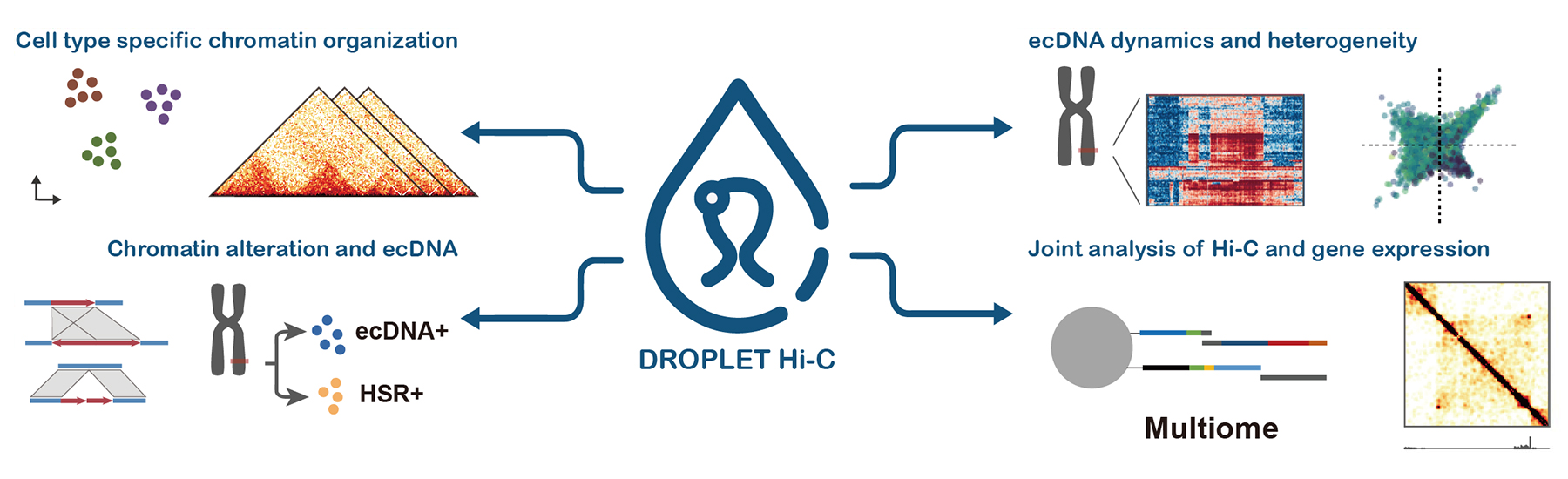
But traditional Hi-C techniques can only give an average view of chromatin organization across thousands or millions of cells. This limits our ability to study how DNA is folded in individual cells, which is crucial for understanding diseases like cancer, where cell types differ significantly. Single-cell Hi-C has emerged as a stopgap solution, but current methods have limitations: they're too expensive and too slow to capture enough cells to be useful.
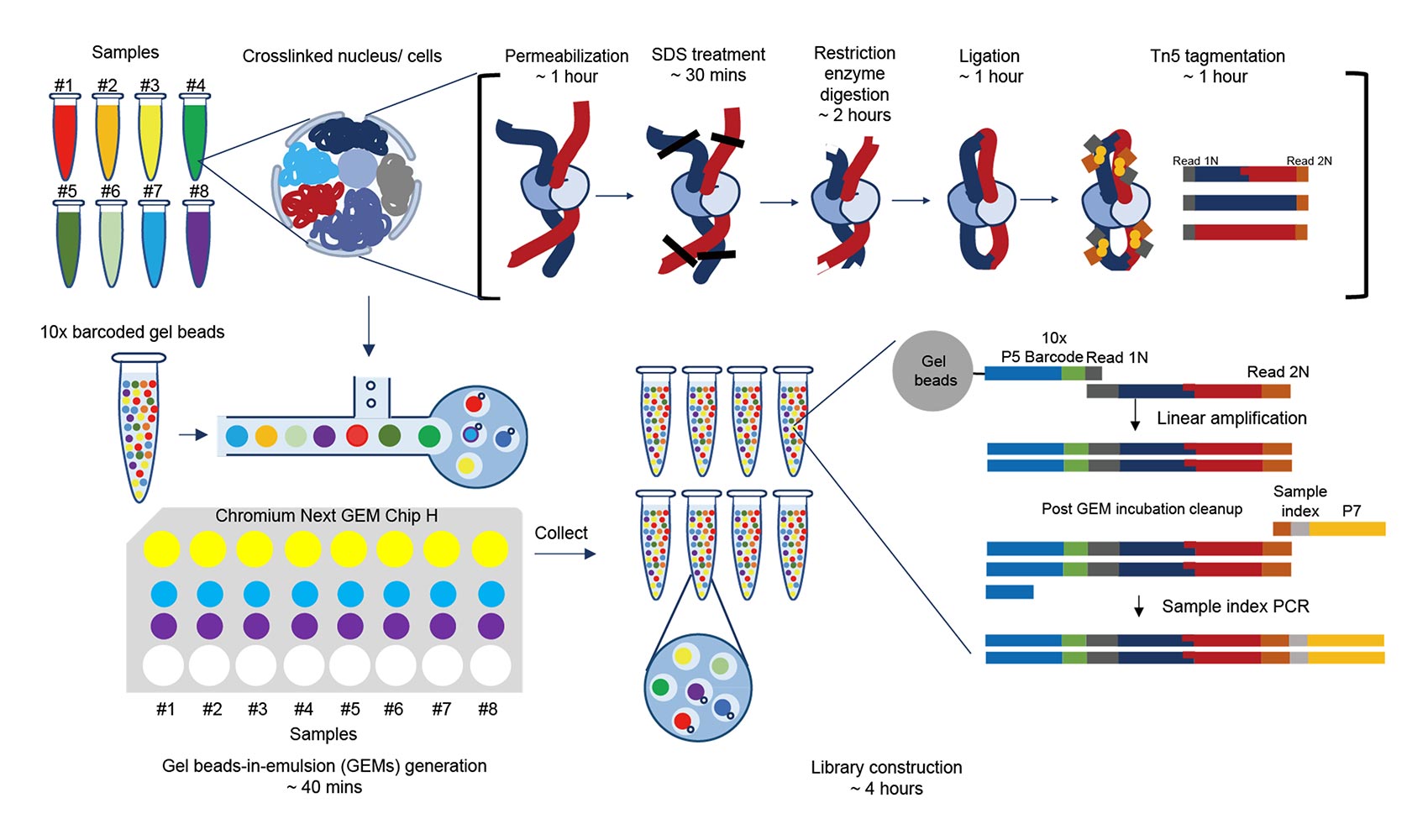
Droplet Hi-C, developed by researchers at the UC San DIego Center for Epigenomics (C4E), is a new method that overcomes these limitations. Recently featured in Nature Biotechnology, it offers a faster, more affordable way to study chromatin architecture in individual cells. Droplet Hi-C uses a high-throughput technology where individual cells are captured inside tiny droplets — think of it like trapping a single cell inside a bubble. Scientists can profile thousands of cells at once this way, making the process much more scalable and efficient.
Droplet Hi-C in Action: Mapping the Mouse Brain
To determine the potential of Droplet Hi-C, researchers turned their attention to the mouse brain, specifically the cortex — a complex region made up of many different types of brain cells. These cells have unique patterns of gene activity, and their chromatin organization reflects that. By using Droplet Hi-C, scientists were able to map the differential chromatin architecture among various cell types in the mouse cortex.
Each of these cell types has distinct chromatin interactions, and understanding these interactions helps scientists determine how genes are regulated differently in each cell type. This is especially useful in understanding brain disorders, where gene regulation often goes awry.
Cancer Research and the Power of Droplet Hi-C
Beyond brain research, Droplet Hi-C is proving to be a powerful tool for cancer research. Tumors are made up of a mix of different cell types, including cancer cells, immune cells, and other supportive cells. To effectively target cancer, researchers need to understand how chromatin is organized in the cancerous cells themselves.
Using Droplet Hi-C, scientists have been able to map the chromatin structure of cancer cells in different types of tumors, including brain cancer (glioblastoma) and blood cancers. Not only does this method reveal how chromatin is folded, but it also helps detect important genetic changes such as “copy number variations” (where parts of the genome are duplicated or missing) and “structural variations” (such as pieces of DNA being rearranged).
One particularly exciting discovery was related to extra-chromosomal DNA (ecDNA), which is a type of DNA that exists outside the normal chromosomes. EcDNA often contains cancer-causing genes (oncogenes) and plays a major role in cancer progression and resistance to treatment. By studying cancer cells using Droplet Hi-C, the team at C4E has gained new insights into how ecDNA evolve and adapt, especially when exposed to treatments like chemotherapy.
Combining Chromatin and Gene Expression Data
One of the most exciting aspects of Droplet Hi-C is that it allows for multi-modal profiling – in other words, it can be combined with other types of data to give a fuller picture of what’s happening in the cell. Researchers can simultaneously study both the 3D structure of the genome and the transcriptome with modified Droplet Hi-C (named Paired Hi-C), which tells us which genes are being actively used by the cell.
This combined approach is especially important in diseases such as cancer, where changes in chromatin architecture often lead to changes in gene expression. By linking the two, scientists can see not just how DNA is folded, but how those folds influence which genes are turned on or off. This could help pinpoint new drug targets or explain why certain cancers are resistant to treatment.
What’s Next for Droplet Hi-C?
With Droplet Hi-C ability to map chromatin architecture at the single-cell level, it opens the door to a wide range of discoveries in fields such as neurobiology, cancer research, and developmental biology. The scalability of the technique means that it can be applied to large, complex tissues, helping researchers uncover new insights into how genes are regulated in health and disease.
In the future, we expect Droplet Hi-C to be used in clinical settings, allowing doctors to profile chromatin organization in patient samples. This could provide personalized insights into how diseases such as cancer progress and how best to treat them. By continuing to refine and expand the applications of Droplet Hi-C, we are just beginning to unlock the full potential of understanding the 3D genome.
