Researcher Stories
- Lungmap 2.0
- Type 2 Diabetes
- Interpreting molecular disease mechanisms
- Roadmap for understanding heart disease
- Computational challenges
- Understanding of the human brain
“This project was a tour de force involving a lot of players in the Center,” said Center Director Dr. Bing Ren. “We had to break new grounds in terms of processing different types of tissues and overcoming not only technical but analytical challenges. We had to develop a lot of new tools,” Ren explained.
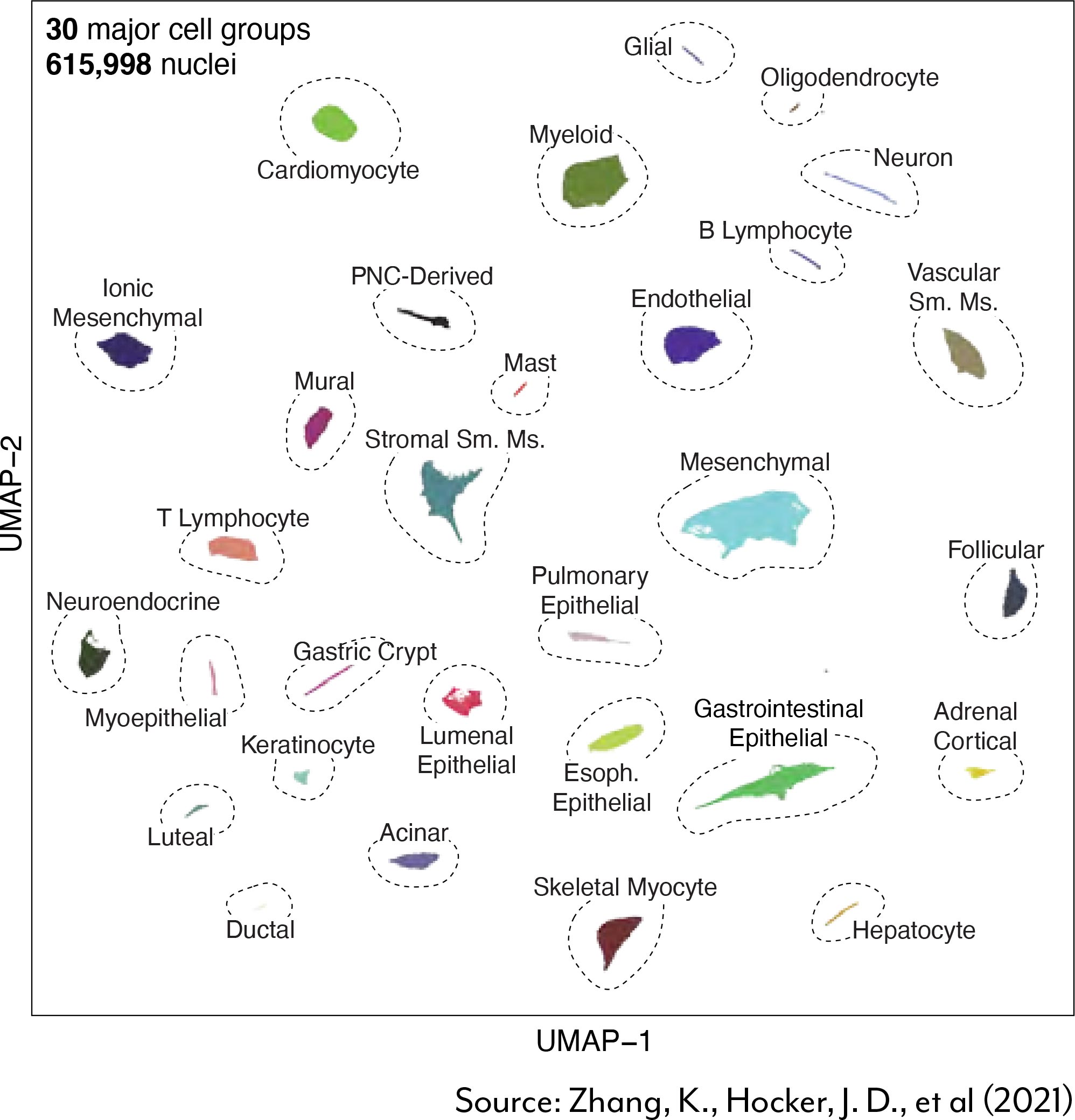
main challenge was defining and standardizing a set of experimental conditions for each tissue type. Clearing this hurdle was a tall order for such a diverse set of sample types, particularly given each tissue’s unique makeup and sensitivity to dissociation. “One of the biggest outcomes for this project, at least internally, was the standardization of the optimization strategy,” said Center Associate Director Dr. Allen Wang. Once they had the process up and running, it took about a week to go from a new tissue to a set of workable conditions, according to Dr. Sebastian Preissl, Associate Director of the Center’s Single-Cell Epigenomic Platform.
“The beauty of this study is that everything was standardized,” Preissl said. “This allowed us to compare cell types that reside in multiple tissues, like fibroblasts.” We can now ask,“How is a fibroblast in the heart different from one in the lung?” With the comprehensive dataset in hand, the Center discovered that, while all fibroblasts shared a core regulatory program, each subtype also possessed tissue-specific regulatory elements associated with various tissue functions.
Another key outcome of the study was the mapping of cCREs in never-before-analyzed cell types. “More than half of the cell types in this atlas were not analyzed or characterized in prior studies,” Ren said. As such, the atlas provides a key resource for investigators interested in interpreting genetic disease variants in these less studied cell types. The Center has already leveraged the map for this very purpose.
Although the map is among the largest human cCRE surveys to date, it is still only an early draft. “We consider this a pilot study,” said Ren. Wang concurred, adding that future versions could include higher cell-type resolution, orthogonal information obtained from other modalities and technologies, and a broader representation of different developmental and disease states.
- Dr. Bing Ren